DNA extraction and testing workshop
December in the UK can be a bit grim: there’s barely any daylight, and everyone is either on holiday or wishes they were. In other words, it’s the perfect time to stay indoors and do a spot of DNA testing, so that’s what the London Hackspace did, holding a public DNA extraction and testing workshop on December 17th.
A DNA testing experiment is a great introduction to molecular biology. It’s fairly robust, so you don’t need a particularly aseptic environment (although it helps); the reagents you will use are cheap and easy to find; and it’s a fun way to experiment on yourself without drinking a petri dishful of H. pylori. The idea is to identify single-nucleotide polymorphisms (the smallest possible genetic variation) in our own DNA.
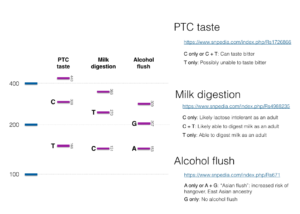
We used SNPedia to help us come up with some interesting SNPs to test. We settled on three for this workshop:
- PTC taste: People with this SNP are unable to taste phenylthiocarbamide and related compounds. Compounds related to phenylthiocarbamide occur in cabbage and raw broccoli and make these foods taste bitter to some people.
- Milk digestion: This is an SNP which causes lactose intolerance.
- Alcohol flush: This well-known SNP, more common in people with East Asian ancestry, affects alcohol metabolism. People with the SNP experience upper body (face and neck) flushing after drinking alcohol and have increased risk of hangover.
On the day of the workshop, after introductions and an quick overview, we got straight into DNA extraction. We used Carolina Biological’s extraction protocol to get template DNA. In our experience getting a good extraction is the hardest part of this entire process (and very frustrating if it doesn’t work out, since you don’t know if it worked until you complete the experiment!), so it’s worth practising with some “known good” primers. We used primers for 28S to make sure our extractions were working well prior to the workshop.
The next stage of the protocol is to do PCR. This (quite amazing) process uses a DNA polymerase (replicating enzyme) to replicate, millions of times, the part of the DNA we are interested in — i.e. the portion of template DNA which surrounds the SNP. The technical details of this part: we used the tetra primer protocol with primers found using BatchPrimer3. We used a 25ul reaction volume consisting of 1ul of template and 1ul of each primer (so 4ul total primer), 12.5ul Taq master mix, and 7.5ul of water.
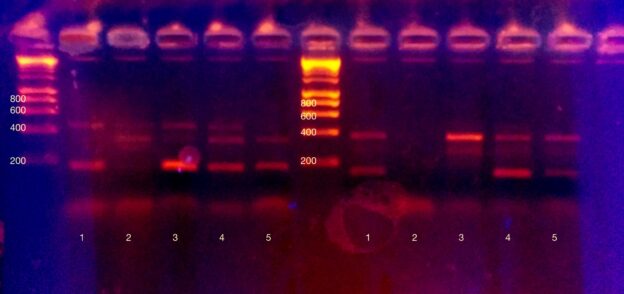
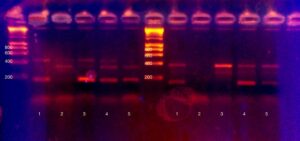
After PCR is the moment of truth. Did it work? Who is likely lactose intolerant? Tensions ran high as we pipetted PCR product onto an agarose gel for electrophoresis. Technical details: we added 8ul of dye to each PCR product, and added 15ul total volume per well. Electrophoresis was at 120V for approximately half an hour. Visualisation was with ethidium bromide and a 300nm (shielded!) light source.
Very excitingly, everyone got at least one working result, and most people got all three results.
This was a really fun workshop. It was a group effort from the Biohackspace, with Ilya designing the primers, Bethan presenting the workshop, Sam providing advice, and Nicholas doing labwork.







This is awesome guys, I wish I attended! I am assuming the electrophoresis and centrifuge are available for the public to use right?